TK6 Mutants Consortium
Human lymphoblastoid TK6 cell line is one of standard cell lines used in in vitro mammalian cells genotoxicity tests. TK6 was originally developed for the thymidine kinase (TK) gene mutation assay. It is now applied to the HPRT gene mutation assay, chromosome aberration tests, micronucleus test, comet assay, sister chromatid exchange test, microarray analysis, GreenScreen HC assay. The high efficiency of cell proliferation and colony formation, karyotype stability, and easily handling of the cells enable to conduct these assays with high performance. TK6 cells are human origin and express wild-type p53 protein. These properties could suggest that the genotoxic and cytotoxic response of TK6 cells against chemical substances are more relevant to intact human cells than that of other mammalian cells. It could be advantageous to consider human hazard and risk. TK6 cells are also useful to understand the mechanism of DNA repair and recombination. The TK gene mutation assay can detect not only point mutations, but also large deletions and recombination because of heterozygosity of the TK gene. The molecular and cytogenetic analysis for the TK mutants had been established, and tells us the characteristic mutational spectrum and repair mechanism response to mutagenic agents. Thus, TK6 cells has great advantage in genotoxicity tests as well as in basic research for DNA repair, mutagenesis, and genomic stability.
The advanced technologies in molecular biology give us new possibility in genotoxicity tests and basic research. Genome editing technologies including CRIPER/Cas9, TALEN, and ZFN can easily alter the genome of any cells. Using this technology, various kind of mutant cells responsible for genes of DNA repair, replication, recombination, and others associated with genome integrity have been developed from TK6 cells. Some of TK6 mutant cells have phenotypically characteristic behaviors showing sensitive or tolerant to a specific mutagen. These properties are useful to understand the DNA repair mechanisms for the mutagens and lead to development a new genotoxicity test systems by combination of the mutant cells.
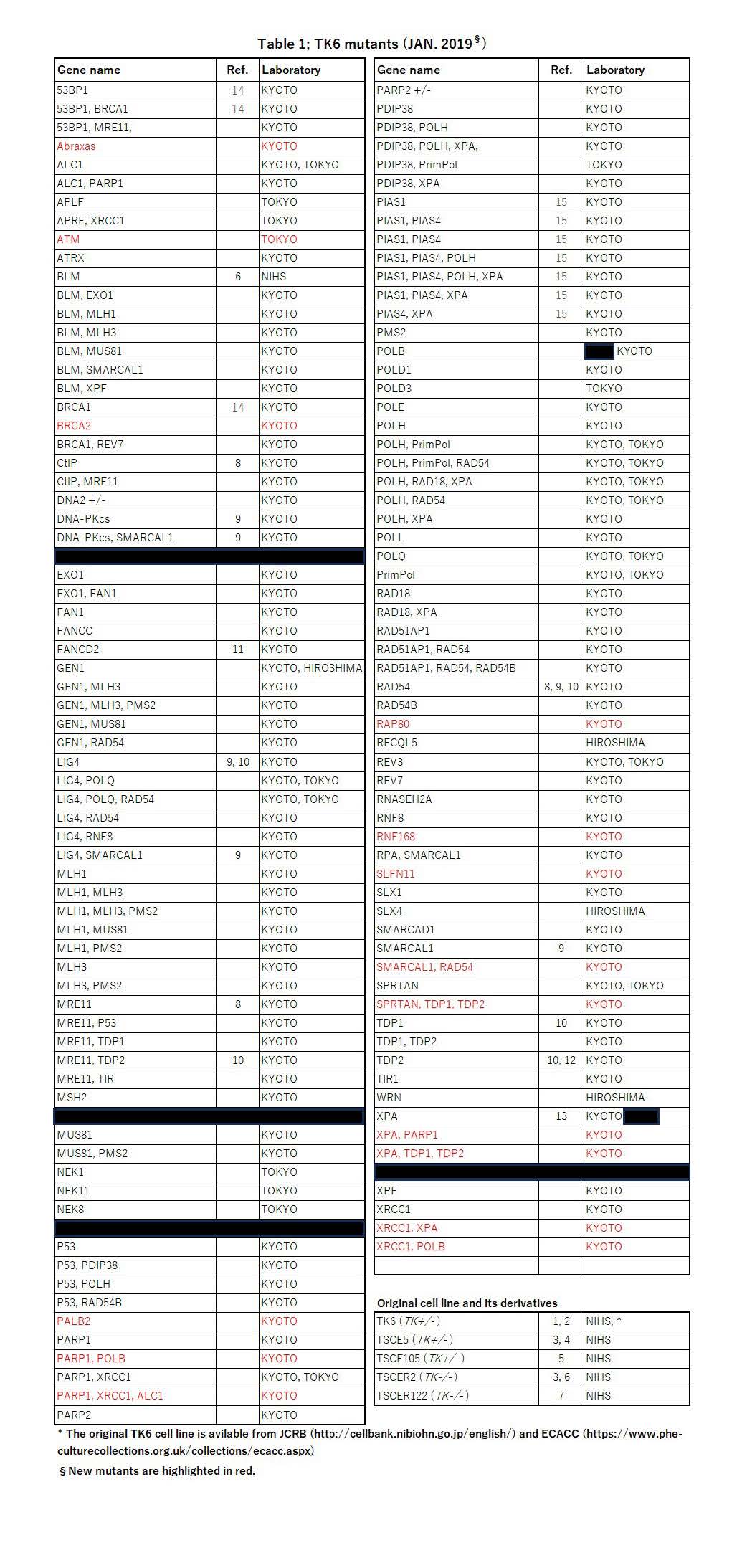
To systematically develop these TK6 mutants and exchange the mutant cells for research works, we organize a consortium as described below. Laboratories from National institute of Health Science (NIHS), Kyoto University (KYOTO), Tokyo Metropolitan University (TOKYO), and Hiroshima University (HIROSHIMA) are currently participating the consortium. We have developed nearly 135 kinds of TK6 mutant cells (Table 1) by knock-out or knock-in of the specific genes using the genome editing technologies. These mutants are originated from parental TK6 cell line or its derivatives. Some research works using the mutants have been published.
If you are interested in the mutant cells, please contact to the contact person(*) in each laboratory and ask the possibility for the supply of the mutant cells or the research collaboration. Some mutant cells may be impossible to be supplied before the publications. Hopefully, these mutant cells will be maintained in a public Cell Bank near future, and supplied freely. We wish that these mutant cells will be useful for your research works.
Consortium Labs
National Institute of Health Scieces (NIHS)
Masamitsu Honma
Masataka Tsuda*
Kyoto University (KYOTO)
Shunichi Takeda*
Hiroyuki Sasanuma
Tokyo Metropolitan University (TOKYO)
Kouji Hirota*
Hiroshima University(HIROSHIMA)
Hiroyuki Kamiya
Tetsuya Suzuki*
Masataka Tsuda
Division of Genome Safety Science,
National Institute of Health Sciences
References
1. Honma M. Generation of loss of heterozygosity and its dependency on p53 status in human lymphoblastoid cells.Environ Mol Mutagen. 2005 Mar-Apr;45(2-3):162-76. PubMed PMID: 15688360.
2. Lorge E, Moore MM, Clements J, O'Donovan M, Fellows MD, Honma M, Kohara A, Galloway S, Armstrong MJ, Thybaud V, Gollapudi B, Aardema MJ, Tanir JY. Standardized cell sources and recommendations for good cell culture practices in genotoxicity testing. Mutat Res. 2016 Oct; 809:1-15. doi: 10.1016/j.mrgentox.2016.08.001. PubMed PMID: 27692294.
3. Honma M, Izumi M, Sakuraba M, Tadokoro S, Sakamoto H, Wang W, Yatagai F, Hayashi M. Deletion, rearrangement, and gene conversion; genetic consequences of chromosomal double-strand breaks in human cells. Environ Mol Mutagen. 2003;42(4):288-98. PubMed PMID: 14673874.
4. Takashima Y, Sakuraba M, Koizumi T, Sakamoto H, Hayashi M, Honma M. Dependence of DNA double strand break repair pathways on cell cycle phase in human lymphoblastoid cells. Environ Mol Mutagen. 2009 Dec;50(9):815-22. doi: 10.1002/em.20481. PubMed PMID: 19402155.
5. Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair (Amst). 2007 Jun 1;6(6):781-8. PubMed PMID: 17296333.
6. Suzuki T, Yasui M, Honma M. Mutator Phenotype and DNA Double-Strand Break Repair in BLM Helicase-Deficient Human Cells. Mol Cell Biol. 2016 Nov 14;36(23):2877-2889. PubMed PMID: 27601585; PubMed Central PMCID: PMC5108877.
7. Yasui M, Kanemaru Y, Kamoshita N, Suzuki T, Arakawa T, Honma M. Tracing the fates of site-specifically introduced DNA adducts in the human genome. DNA Repair (Amst). 2014 Mar; 15:11-20.doi: 10.1016/j.dnarep.2014.01.003. PubMed PMID: 24559511.
8. Hoa NN, Akagawa R, Yamasaki T, Hirota K, Sasa K, Natsume T, Kobayashi J, Sakuma T, Yamamoto T, Komatsu K, Kanemaki MT, Pommier Y, Takeda S, Sasanuma H. Relative contribution of four nucleases, CtIP, Dna2, Exo1 and Mre11, to the initial step of DNA double-strand break repair by homologous recombination in both the chicken DT40 and human TK6 cell lines. Genes Cells. 2015 Dec;20(12):1059-76. doi: 10.1111/gtc.12310. PubMed PMID: 26525166.
9. Keka IS, Mohiuddin, Maede Y, Rahman MM, Sakuma T, Honma M, Yamamoto T, Takeda S, Sasanuma H. Smarcal1 promotes double-strand-break repair by nonhomologous end-joining. Nucleic Acids Res. 2015 Jul 27;43(13):6359-72. doi:10.1093/nar/gkv621. PubMed PMID: 26089390; PubMed Central PMCID: PMC4513880.
10. Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, Akter S, Tsuda M, Furuta R, Tsusui K, Takeda S, Sasanuma H. Mre11 Is Essential for the Removal of Lethal Topoisomerase 2 Covalent Cleavage Complexes. Mol Cell. 2016 Nov 3;64(3):580-592.doi: 10.1016/j.molcel.2016.10.011. PubMed PMID: 27814490.
11. Hashimoto K, Sharma V, Sasanuma H, Tian X, Takata M, Takeda S, Swenberg JA, Nakamura J. Poor recognition of O6-isopropyl dG by MGMT triggers double strand break-mediated cell death and micronucleus induction in FANC-deficient cells. Oncotarget. 2016 Sep 13;7(37):59795-59808. doi: 10.18632/oncotarget.10928. PubMed PMID: 27486975.
12. Marchand C, Abdelmalak M, Kankanala J, Huang SY, Kiselev E, Fesen K, Kurahashi K, Sasanuma H, Takeda S, Aihara H, Wang Z, Pommier Y. Deazaflavin Inhibitors of Tyrosyl-DNA Phosphodiesterase 2 (TDP2) Specific for the Human Enzyme and Active against Cellular TDP2. ACS Chem Biol. 2016 Jul 15;11(7):1925-33. doi: 10.1021/acschembio.5b01047. PMID:27128689
13. Sassa A, Kamoshita N, Kanemaru Y, Honma M, Yasui M. Xeroderma Pigmentosum Group A Suppresses Mutagenesis Caused by Clustered Oxidative DNA Adducts in the Human Genome. PLoS One. 2015 Nov 11;10(11):e0142218. doi: 10.1371/journal.pone.0142218. PMID:26559182
14. Sasanuma H, Tsuda M, Morimoto S, Saha LK, Rahman MM, Kiyooka Y, Fujiike H, Cherniack AD, Itou J, Callen Moreu E, Toi M, Nakada S, Tanaka H, Tsutsui K, Yamada S, Nussenzweig A, Takeda S. BRCA1 ensures genome integrity by eliminating estrogen-induced pathological topoisomerase II-DNA complexes. Proc Natl Acad Sci U S A. 2018 Nov 6;115(45):E10642-E10651. doi: 10.1073/pnas.1803177115. PMID:30352856
15. Mohiuddin M, Evans TJ, Rahman MM, Keka IS, Tsuda M, Sasanuma H, Takeda S. SUMOylation of PCNA by PIAS1 and PIAS4 promotes template switch in the chicken and human B cell lines. Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):12793-12798. doi: 10.1073/pnas.1716349115. PMID:30487218