AMES/QSAR International Collaborative Study
Robust Quantitative Structure–Activity Relationship (QSAR) models defining toxicological endpoints are desirable to enable regulatory authorities to identify chemicals possibly causing adverse effects without performing actual toxicological studies. Much effort has been invested in the development of QSAR models to predict Ames mutagenicity, among many toxicological endpoints, to exploit the large body of Ames data and the strong correlation between chemical structure and Ames mutagenicity. Ames results are important for decisions on the development of chemical products and pharmaceuticals and the assessment of chemical safety, given that a positive result corresponds to increased cancer risk from exposure to the chemical even at a low level. The ICH-M7 guideline (Assessment and control of DNA-reactive impurities in pharmaceuticals to limit potential carcinogenic risk) currently recommends two QSAR models (expert rule-based and statistical) to predict Ames mutagenicity for initially assessing DNA-reactive impurities in pharmaceuticals. This is the first international guideline addressing the use of QSAR in lieu of an actual toxicological study for human health assessment. Thus, QSAR models for Ames mutagenicity now require much greater prediction power to ensure the safety of chemicals. To increase this prediction power, experimental data sets as training data to build the models are important. Large numbers of highly reliable data sets will allow development and improvement of QSAR models with high predictive power.
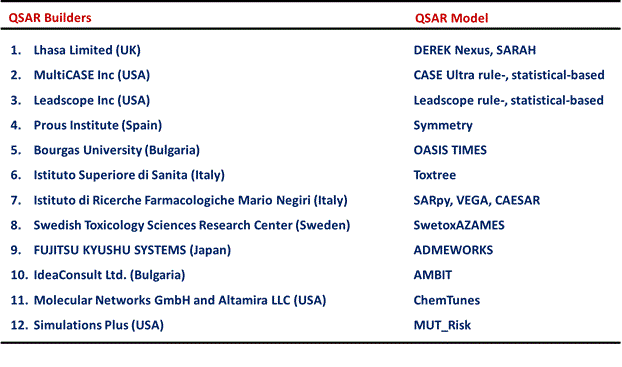
The Division of Genetics and Mutagenesis, National Institute of Health Sciences (DGM/NIHS) has Ames mutagenicity data for approximately 12,000 new chemicals. The Ames assays were conducted according to the OECD TG471 guideline and Industrial Safety and Health Act in Japan under GLP-compliant conditions. We now provide these Ames data to QSAR builders/vendors to improve their QSAR models for predicting Ames mutagenicity with the permission of the Industrial Safety and Health Department of the Ministry of Health, Labor and Welfare (MHLW), Japan. The Ames/QSAR international collaborative study leaded by DGM/NIHS launched on 2014. Because most of the Ames data are confidential, the QSAR builders/vendors participating in the project must execute a confidentiality agreement. Twelve QSAR builders/vendors from USA, UK, Italy, Spain, Bulgaria, Sweden, and Japan are currently participating in this project (Table 1).
Three phase trials were designed to allow testing of the hypothesis that the knowledgebase expansion enhances prediction results. DGM/NIHS provides about 4,000 chemicals in each trial without their Ames results to the QSAR builders/vendors, who calculated the Ames mutagenicity from their QSAR models. DGM/NIHS evaluated the performance of the QSAR models (sensitivity, specificity, and other criteria) and disclosed the Ames results. The Ames results are classified into three categories. Class A is strongly positive, in which the chemical induces more than 1,000 revertants/mg in at least one Ames strain in the presence or absence of rat S9. Class B is positive but not in class A. Class C is negative, in which all of Ames strains exhibit negative responses in both the presence and absence of rat S9. The QSAR builders/vendors integrated the Ames results as knowledges into their QSAR models as training data and entered the next trial. Finally, all of the QSAR models will be much improved in prediction of Ames mutagenicity. We believe that the outcome of this project will be of great benefit for QSAR builders/vendors, QSAR users, and regulatory authorities.
All trials (phase I, II, III) have been completed recently. We challenged totally 12,140 chemicals (Table 2). After the trial, all QSAR models have considerably improved. Some QSAR models showed nearly 90% prediction power, which is the same level as that of the inter-laboratory correspondence of the Ames assay.
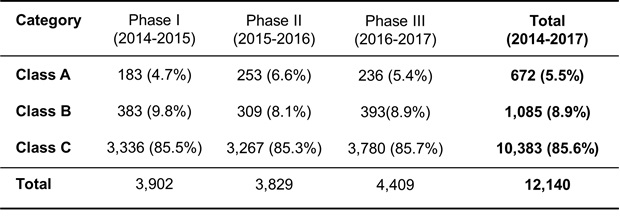
We disclose class A chemicals. These chemicals must have important alert structures strongly associated with Ames mutagenicity. The phase I chemicals (3,902) include 183 class A chemicals. The phase II chemicals (3,829) include 253 class A chemicals. The phase III chemicals (4,409) include 236 class A chemicals.
I talked about the outcome of this project in the 12th International Conference on Environmental Mutagen (ICEM2017) in Inchon, Korea on November 12-16, 2017 (http://www.icem2017.org/) as plenary lecture. The presentation file is available here.
The outcome of this project has been published in the Special Issue of Mutagenesis (Vol. 34, No.1, 2019) “In silico approaches to genetic toxicology”. The paper is available here.
If you have any question about this project, please contact with honma@nihs.go.jp
Masamitsu Honma
Division of Genetics and Mutagenesis,
National Institute of Health Sciences
Last update; June 2019